RNA and protein modifications
RNA and protein modifications
Most RNAs and proteins involved in mRNA maturation and translation are post-transcriptionally or post-translationally modified by numerous chemical groups, which improve the efficiency, fidelity and regulation of fundamental biological processes such as mRNA maturation, stability or translation.
These modifications are widely conserved throughout evolution, but their role has long been overlooked due to the frequent absence of obvious phenotypes in unicellular organisms, or to the still unknown nature of the enzymes (or writers) that add these marks. Over the last decade, numerous studies have demonstrated the critical role of these modifications. Writers now appear to be essential for several cellular processes, as well as for human pathologies such as cancers and neurodevelopmental disorders.
The team is seeking to decipher the role of these post-transcriptional and post-translational modifications in central biological processes (mRNA maturation, translation and decay), cell proliferation and organ development, and to describe the consequences of pathogenic mutants on the functions of the enzymes responsible for these modifications.
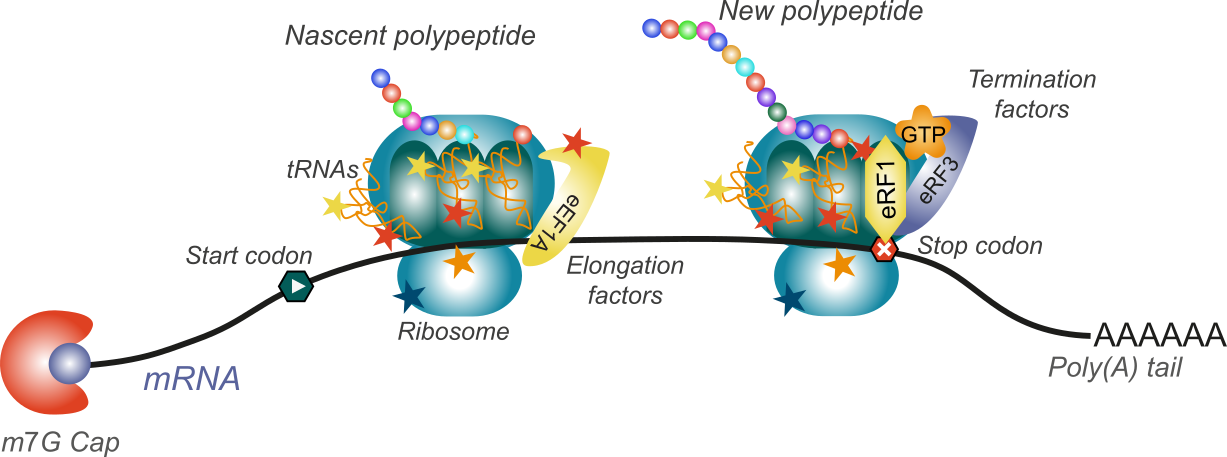
RNA and protein modifications are central for protein synthesis
(Artwork : Morgane Corre)
The team uses a wide range of experimental approaches, made possible by the local infrastructure set up at BIOC, for molecular biology, protein expression and purification, structural biology, cell biology, RNA biochemistry...
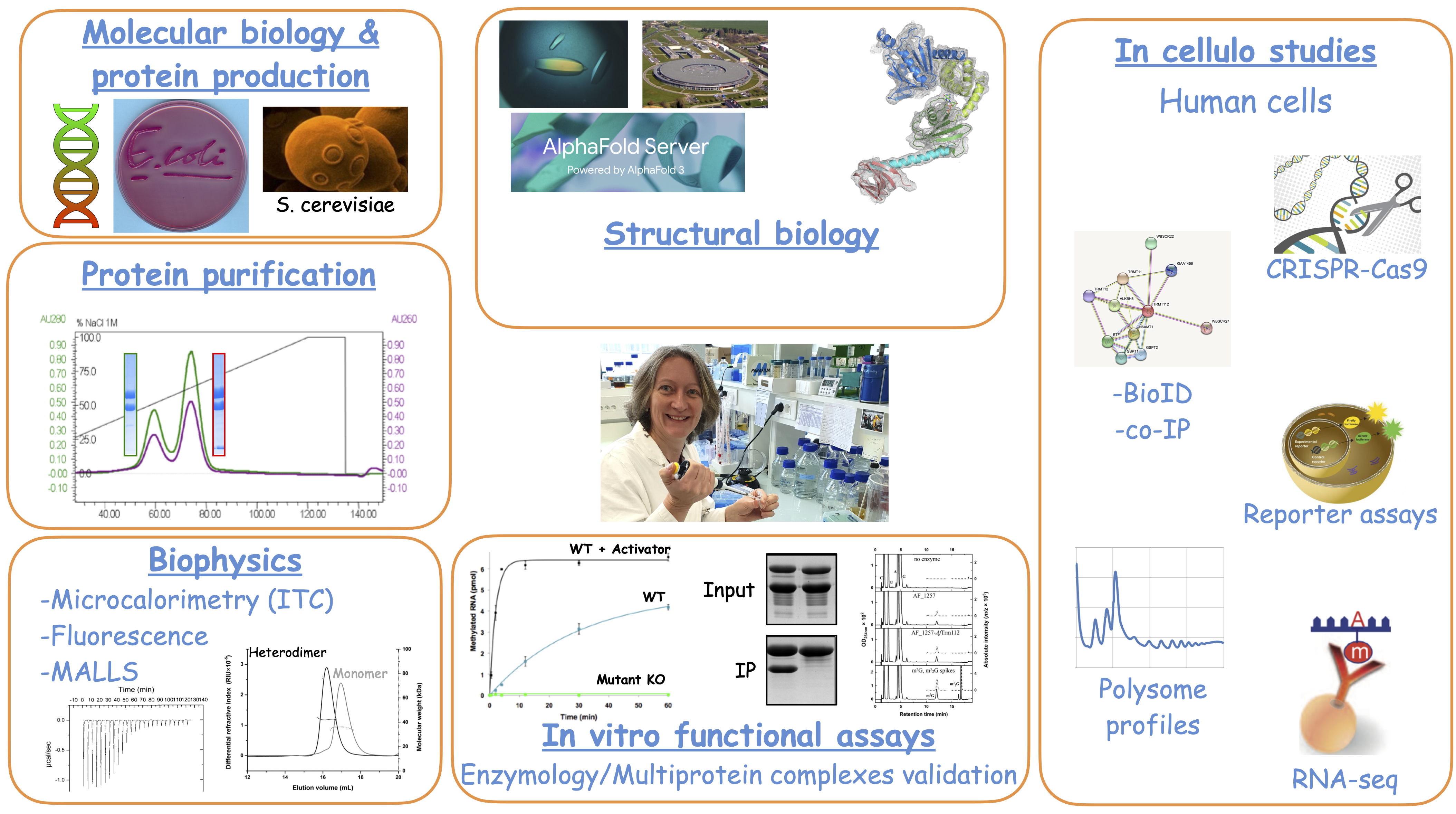
Some techniques used in the lab
TRMT112 protein, a major activator of methyltransferases
Since many years , our team is studying the TRMT112 protein, which is present in all three domains of life and acts as an allosteric regulator of several methyltransferases (MTases). We are particularly interested in the TRMT112 protein from eukaryotic organisms, which interacts with MTases modifying factors (tRNA, rRNA, proteins...) involved in the maturation and translation processes of messenger RNAs (mRNAs) (Wang et al; 2024)
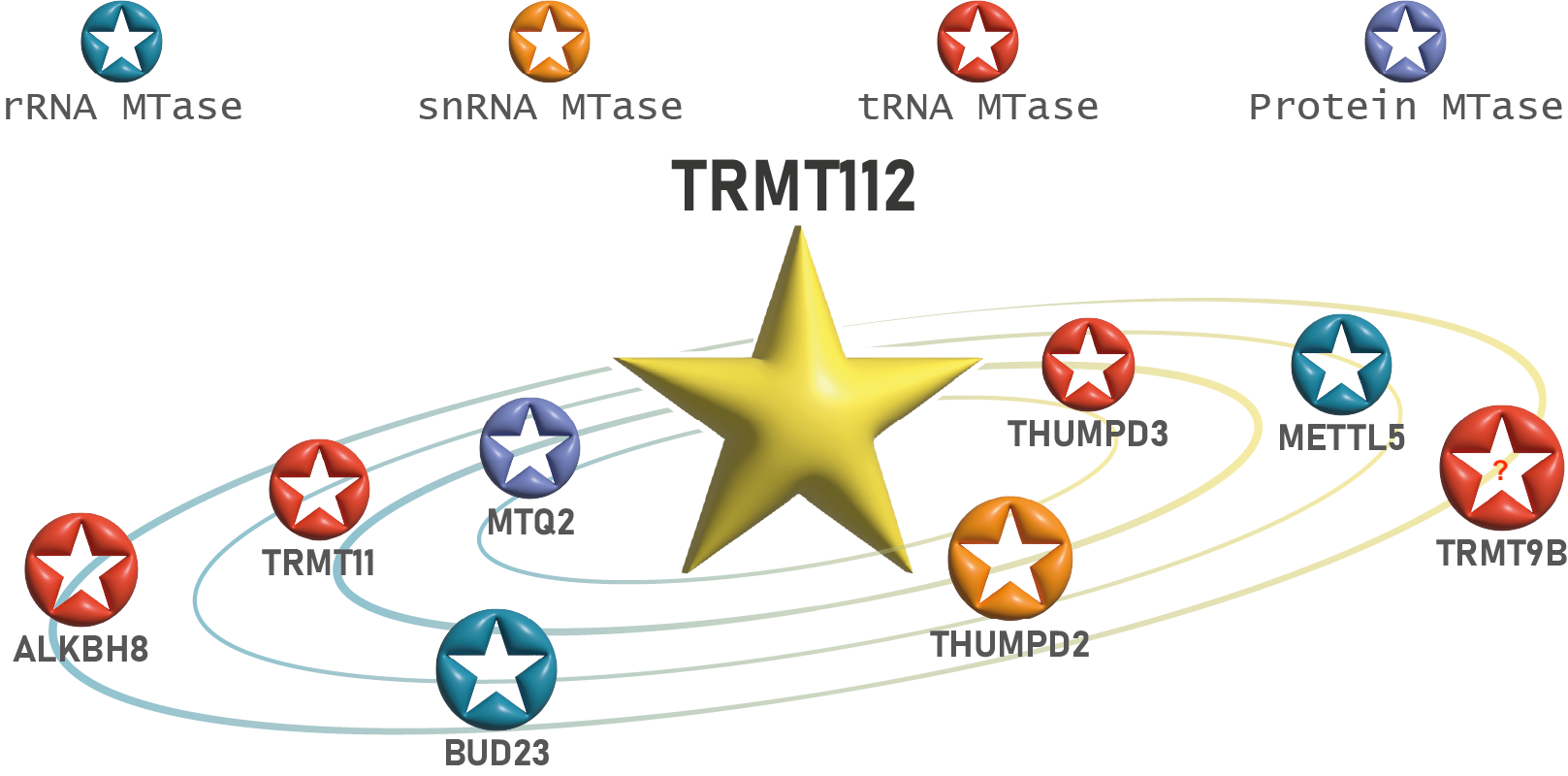
Human TRMT112 interaction network
(Artwork : Morgane Corre)
In humans, the TRMT112-BUD23 and TRMT112-METTL5 complexes catalyze the formation of N7-methylguanosine (m7G) and N6-methyladenosine (m6A) on 18S rRNA, respectively, and participate in the 40S ribosome subunit biogenesis pathway. The TRMT112-MTQ2 complex modifies the translation termination factor eRF1, which is essential for the release of newly synthesized proteins. The TRMT112-ALKBH8, TRMT112-THUMPD3 and TRMT112-TRMT11 complexes contribute to translation elongation by modifying tRNAs. The TRMT112-THUMPD2 complex modifies small nuclear RNA U6 (snRNA U6), which is involved in pre-messenger RNA splicing. Finally, the precise molecular and biological functions of the TRMT112-TRMT9B complex remain to be elucidated. Some of these complexes are also present in certain archaea, underscoring the strong similarities between the protein synthesis machineries of eukaryotes and archaea.
Using a wide range of approaches and with the help of various collaborators, the team has studied the interaction network of human, Saccharomyces cerevisiae yeast and archaeal TRMT112 proteins with different MTases. We discovered the biochemical functions of the TRMT112-METTL5, TRMT112-THUMPD3 and TRMT112-THUMPD2 complexes (Tran et al.; Nucleic Acids Research, 2019 / Wang et al.; Nucleic Acids Research, 2023). We also characterized the enzymatic activities of all these complexes and obtained structural information on several of them. This revealed that these different MTases interact in a very similar mode with TRMT112 despite low sequence identity (less than 20%), and are therefore in direct competition with each other. These studies made it possible to clarify the molecular mechanisms used by TRMT112 to activate these MTases. TRMT112 stabilizes these MTases in vivo and in vitro, confers them with the capacity to bind S-adenosyl-L-methionine (SAM), the methyl group donor used by all these MTases, and also contributes directly to substrate binding by certain complexes.
As deregulation of expression levels of TRMT112 protein or associated MTases affects cancer cell proliferation, and as pathogenic mutations associated with neurodevelopmental disorders have been identified in several MTases interacting with TRMT112, the team aims to uncover the biological functions of these MTases and their importance in nervous system development.



