Bioinformatique structurale
BIOS Research Group BIOcomputing and Structure
We are a part of the Laboratoire de Biologie Structurale de la Cellule in the Biology Department of
the Ecole Polytechnique and the Centre National de la Recherche Scientifique.
We are located in the Ecole Polytechnique, Palaiseau, just outside of Paris.
snail-mail: Laboratoire de Biologie Structurale de la Cellule, Ecole Polytechnique, 91128 Palaiseau, France.
People

Thomas Simonson, Senior Research Scientist, CNRS
Thomas Gaillard, Lecturer, Ecole Polytechnique
David Mignon, software engineer; Xingyu Chen, postdoc
Ivan Reveguk, Anna Mazurek, Ph.D. students
Research interests
Our group uses computer modeling and simulation to study and to engineer structure-function relationships in biomolecules. We use state-of-the-art simulation techniques, including computational protein design, molecular dynamics, continuum electrostatics, and free energy perturbation techniques, and a significant portion of our effort is directed at the development of new techniques as new needs arise. We are especially interested in protein engineering, protein-ligand recognition, and the inverse protein folding problem. We have taken part for many years in the development of free energy perturbation techniques for proteins, and these techniques are now maturing into a reliable and remarkable tool. Recent applications to aminoacyl-tRNA synthetases have provided insights into the translation of the genetic code. Protein design using directed evolution is an important research direction in the group. We have developed the Proteus software package for physics-based computational protein design. 
Proteus software for protein design
Other software and data
Some recent publications
A complete list of recent publications is available here. See also: orcid.org/0000-0002-5117-7338
T. Simonson, editor (2021) Methods in Molecular Biology; Springer Protocols, vol 2405, Computational Peptide Science (book, 424 pages)
E. Michael & T. Simonson (2022) Current Opinion in Structural Biology 72, 46-54. How much can physics do for protein design?
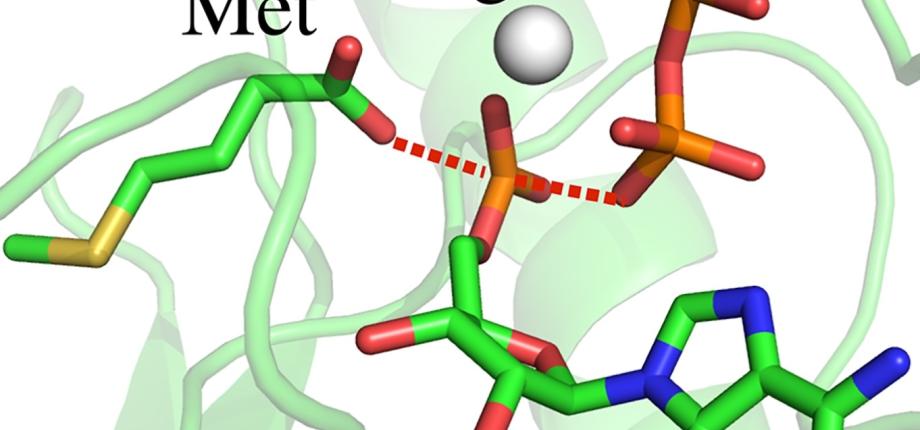
V. Opuu, G. Nigro, T. Gaillard, E. Schmitt, Y. Mechulam & T. Simonson (2020) PLoS Computational Biology, 16, e1007600. Adaptive landscape flattening allows the design of both enzyme:substrate binding and catalytic power.
V. Opuu, G. Nigro, C. Lazennec-Schurdevin, Y. Mechulam, E. Schmitt & T. Simonson (2023) Protein Science, 32, e4738. Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Teaching material
Other interesting links:


